In this tutorial you will learn how to use Exasol UDFs with Python in a machine learning context.
This tutorial uses database objects in the free public demo system hosted by Exasol. To get access to the public demo system, sign up here.
Introduction
In this tutorial you will learn how to test a machine learning model within Exasol by using a user defined function (UDF) written in Python and an already trained model with sample data.
The tutorial will demonstrate that there is no need to export data from Exasol to a different system to apply machine learning models. Everything can be done using UDFs directly within the Exasol database where the data is stored.
To follow the tutorial you should have basic Python programming knowledge and a basic understanding of Exasol and of what UDFs are. The tutorial does not require a deep understanding of data science or machine learning methods.
For an introduction to Exasol, see Get started.
For more information about user defined functions (UDFs), see User defined functions (UDFs).
Model testing with UDFs
A machine learning model is typically designed in multiple steps: load the data -> normalize attributes -> build the model -> train the model -> refine parameters -> evaluate performance. In this tutorial we have already developed an adequate model and trained it accordingly. We will focus on the final two steps, which is running the model on the test data and evaluating its performance.
For a complete end-to-end example of how to apply machine learning techniques in Exasol, visit our Data Science GitHub repository.
Procedure
You can choose to write the UDFs in your SQL client, or you can create them in a Python development environment such as a Jupyter Notebook using the PyExasol package. The advantage of creating the UDFs in a Python IDE is that you can play around with dataframes in Python and visualize the results in the IDE. Both methods are shown in the tutorial, and the result will be the same regardless of which method you choose.
About the data set
This tutorial uses real-world data from the IDA 2016 Industrial Challenge data set. The data was provided by the truck manufacturer Scania and was used in the IDA 2016 challenge to predict which failures were related to a specific component of a truck’s air pressure system (APS) as opposed to failures unrelated to the APS.
The tutorial uses the IDA
schema in the Exasol public demo database. To get access to the public demo, sign up here .
Step 1: Connect to the Exasol public demo system
SQL client
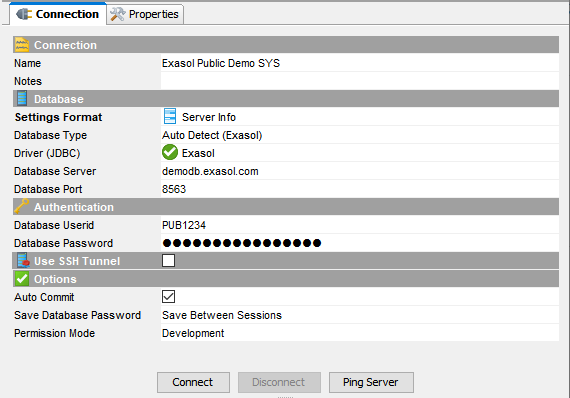
Connect to the demo system using the SQL client of your choice. Enter the credentials that you received when you signed up for the demo.
Python environment
First, install the PyExasol package in your local Python environment. In this tutorial we use Jupyter Notebook, but you can use any Python environment.
When PyExasol is installed in your environment, you can start writing your script by specifying the connection details for the Exasol demo system.
Replace the username and password in the example with the credentials you received when you signed up for the demo. Replace the schema name with your username so that the UDFs are created in your own schema.
EXASOL_HOST = "demodb.exasol.com:8563"
EXASOL_USER = "<YOUR_USER_NAME>"
EXASOL_PASSWORD = "<YOUR_PASSWORD>"
SCHEMA_NAME ="<YOUR_USER_NAME>"Test the connection to the database using the following statement:
import pyexasol
import textwrap
# create the connection
C = pyexasol.connect(dsn=EXASOL_HOST, user=EXASOL_USER, password=EXASOL_PASSWORD, compression=True)
# test the connection
stmt = C.execute("SELECT 42 FROM DUAL")
print(stmt.fetchone()[0])You now have an open connection to the demo system and can move on to the next step - Examine the data.
Step 2: Examine the data
To understand the data structure before you apply the model, look at the relevant table in the IDA schema. Since you will use an already trained model, you only need the test data in a normalized format. This data is stored in the table IDA_TEST_TRANSFORMED.
SQL client
-- count the number of rows in the table
SELECT COUNT(*) FROM IDA.IDA_TEST_TRANSFORMED;
-- get a preview of the test data
SELECT * FROM IDA.IDA_TEST_TRANSFORMED LIMIT 10;
Python environment
# count the number of rows in the table
stmt = C.execute("SELECT COUNT(*) from IDA.IDA_TEST_TRANSFORMED")
print(stmt.fetchone()[0])# get a preview of the test data
df = C.export_to_pandas("SELECT * FROM IDA.IDA_TEST_TRANSFORMED LIMIT 5")
print(df.head())# close the connection
C.close()Step 3: Prepare the environment
When you have set up a connection and you understand the data structure, you can start preparing your environment for creating the UDFs.
Script language containers
A script language container (SLC) in Exasol contains a complete Linux distribution and all libraries that are required to run a specified script language. Exasol ships with a default SLC for Python 3, which can be used to run Python UDFs out of the box in the database. This container includes some commonly used data science modules. If you need additional libraries that are not included in the default container, or if you want to use another version of Python, you can create your own script language containers to get a custom UDF environment tailored to your needs. For more information, see Adding new packages to existing script languages.
BucketFS
Exasol systems use a distributed file system called BucketFS. A BucketFS service contains buckets that can store files. Each bucket can have different access privileges. The Exasol demo system contains a bucket where the model that you will use in this tutorial is stored. In the following step you will define the details to allow the UDF to access the content in this bucket.
SQL client
Use the following command to open your schema on the demo system. Replace <YOUR_USER_NAME> with your username.
NOTE: The schema is only set for the current session. If you disconnect from the demo system and then reconnect, you have to execute this statement again.
-- open your own schema
OPEN SCHEMA <YOUR_USER_NAME>;Python environment
First, you need to define some variables that describe where in the BucketFS the model that we want to test is located.
# bucketfs service name
EXASOL_BUCKETFS_SERVICE = "bucketfs1"
# bucketfs bucket name
EXASOL_BUCKETFS_BUCKET = "demo_ida"
# path to the read-only mounted BucketFS inside the running UDF container
EXASOL_BUCKETFS_PATH = f"/buckets/{EXASOL_BUCKETFS_SERVICE}/{EXASOL_BUCKETFS_BUCKET}"
# boolean enabling https
EXASOL_BUCKETFS_USE_HTTPS = True
# filename of the model
model_file = "classifier3.pkl"Then, use the following command to open your schema on the demo system.
NOTE: The schema is only set for the current session. If you disconnect from the demo system and then reconnect, you have to execute this statement again.
# create connection
C = pyexasol.connect(dsn=EXASOL_HOST, user=EXASOL_USER, password=EXASOL_PASSWORD, compression=True)
# open your own schema
C.execute(f"OPEN SCHEMA {SCHEMA_NAME}")Step 4: Create a script to test the model
In this step you will create a Python UDF script called IDA_TEST_MODEL. This UDF will be used in the following step to predict results for the test data (whether a failure is an APS failure or not).
The input parameters for the UDF are all the columns that should be considered for the failure prediction. The UDF will emit results that are the predicted classes (first column) joined to the transformed test data. By joining the predicted class labels to the test data, we ensure that the predicted and real class labels remain properly ordered/linked for evaluation, since we do not have a smaller set of columns that uniquely identifies each row.
Because we have a large number of attributes to take into account, we will use the system tables that contain the column names and column types to generate the attributes instead of typing them manually.
SQL client
Copy the following script and paste it into your SQL client. Do not execute the script, since the columns have not been included yet.
--/
CREATE OR REPLACE PYTHON3 SET SCRIPT IDA_TEST_MODEL(CLASSIFIER_PATH VARCHAR(200),<list of columns>
)
EMITS (CLASS_PRED DECIMAL,<list of columns>
) AS
import pandas as pd
from sklearn.externals import joblib
# Test classifier
def test(X, class_col_name, model_path=None):
# Convert columns to numeric data types
X = X.apply(pd.to_numeric)
X_data = X.loc[:, X.columns != class_col_name]
# Load model from EXABucket
clf = joblib.load(model_path)
# Predict classes of test data
return clf.predict(X_data)
def run(ctx):
# Input argument
num_non_data_cols = 1
classifier_path = ctx.CLASSIFIER_PATH
df = ctx.get_dataframe(num_rows='all', start_col=num_non_data_cols)
# Test the classifier
y_pred = test(df, class_col_name='CLASS', model_path=classifier_path)
# Add class predictions as first column of test DataFrame
df_pred = (pd.DataFrame(y_pred, columns=['CLASS_PRED'])).join(df)
# Convert columns to numeric data types
df_pred = df_pred.apply(pd.to_numeric)
# Output data
ctx.emit(df_pred)
/Execute the following small helper statement to produce a list of columns to be included in the script:
SELECT group_concat(column_name || ' ' || column_type
ORDER BY column_ordinal_position)
AS input_cols
FROM exa_all_columns c
WHERE column_schema = 'IDA'
AND column_table = 'IDA_TEST_TRANSFORMED';Copy the output and paste it into the script where the placeholders <list of columns> are.
Execute the script to generate the UDF.
If you are using DbVisualizer, you must disable Parameterized SQL
before executing a script containing &
characters. Otherwise, the client will request that you enter values for the parameters. For more information, see the DbVisualizer documentation.
Python environment
# read one line of the data set to get the structure
df = C.export_to_pandas("SELECT * FROM IDA.IDA_TEST_TRANSFORMED LIMIT 1")
# create tables for data
column_names = list(df.columns)
###column_types = ["VARCHAR(3)"] + ["DECIMAL(18,2)"] * (len(column_names))
###column_desc = [" ".join(t) for t in zip(column_names, column_types)]
# numeric data
column_types = ["INT"] + ["DOUBLE"] * (len(column_names) - 1)
column_desc = [" ".join(t) for t in zip(column_names, column_types)]
# create script output column descriptions
# one class label, numeric data
out_column_types = ["INT"] * 2 + ["DOUBLE"] * (len(column_names))
out_column_desc = [" ".join(t) for t in zip(["class_pred"] + column_names, out_column_types)]
# create script to test the model
sql = textwrap.dedent(f"""\
CREATE OR REPLACE PYTHON3 SET SCRIPT IDA_TEST_MODEL(CLASSIFIER_PATH VARCHAR(200), {", ".join(column_desc)}
)
EMITS ({", ".join(out_column_desc)}) AS
import pandas as pd
from sklearn.externals import joblib
# Test classifier
def test(X, class_col_name, model_path=None):
# Convert columns to numeric data types
X = X.apply(pd.to_numeric)
X_data = X.loc[:, X.columns != class_col_name]
# Load model from EXABucket
clf = joblib.load(model_path)
# Predict classes of test data
return clf.predict(X_data)
def run(ctx):
# Input argument
num_non_data_cols = 1
classifier_path = ctx.CLASSIFIER_PATH
df = ctx.get_dataframe(num_rows='all', start_col=num_non_data_cols)
# Test the classifier
y_pred = test(df, class_col_name='CLASS', model_path=classifier_path)
# Add class predictions as first column of test DataFrame
df_pred = (pd.DataFrame(y_pred, columns=['CLASS_PRED'])).join(df)
# Convert columns to numeric data types
df_pred = df_pred.apply(pd.to_numeric)
# Output data
ctx.emit(df_pred)
/
""")
C.execute(sql)
Step 5: Let the model predict
You are now ready to run the data science model for prediction and evaluation on the data using the script that you just created. The results will be stored in the table TEST_PREDICTIONS.
The UDF TEST_MODEL takes the path to the model and all the input columns as input parameters. The first parameter that is handed over to the UDF is the path to the model in BucketFS. The following input parameters are the columns of the table containing the test data.
SQL client
Copy the following statement into your SQL client. Do not execute the statement, as the columns have not been included yet.
-- create a new table in your schema
CREATE OR REPLACE TABLE IDA_TEST_PREDICTIONS AS
SELECT IDA_TEST_MODEL('/buckets/bucketfs1/demo_ida/classifier3.pkl',<list of columns>)
FROM IDA.IDA_TEST_TRANSFORMED;Execute the following helper statement to generate the list of columns:
SELECT group_concat(column_name
ORDER BY column_ordinal_position)
AS input_cols
FROM exa_all_columns c
WHERE column_schema = 'IDA'
AND column_table = 'IDA_TEST_TRANSFORMED';Copy the output from this statement and paste it into the first statement, replacing the placeholder <list of columns>.
Execute the first statement to generate the table TEST_PREDICTIONS.
Python environment
# test model
sql = textwrap.dedent(f"""\
CREATE OR REPLACE TABLE IDA_TEST_PREDICTIONS AS
SELECT IDA_TEST_MODEL('{EXASOL_BUCKETFS_PATH}/{model_file}', {", ".join(column_names)})
FROM IDA.IDA_TEST_TRANSFORMED
""")
C.execute(sql)The table that you generated in this step will be used in the next step to evaluate the results.
Step 6: Evaluate the model
The result table IDA_TEST_PREDICTIONS contains a column with the prediction made by the model CLASS_PRED and the actual label as the ground of truth in the column CLASS. In this step you will compare these two columns to evaluate how well the model performed.
A cost function called ida_cost() is used here as a performance metric, which implements the cost function specified in the problem description. A confusion matrix that displays how the instances were classified is also generated.
SQL client
Execute the following script:
--/
CREATE OR REPLACE PYTHON3 SET SCRIPT IDA_EVALUATE_RESULTS(CLASS_PRED double, CLASS double)
EMITS (RESULTS varchar(200),PREDICTED_NEG double, PREDICTED_POS double, TOTAL_COSTS double) AS
from sklearn.metrics import confusion_matrix
import pandas as pd
import pyexasol
import numpy as np
# Define cost function from the problem description
def ida_cost(y, y_pred):
false_preds = y - y_pred
num_false_pos = (false_preds < 0).sum()
num_false_neg = (false_preds > 0).sum()
return 10 * num_false_pos + 500 * num_false_neg
def evaluate(df):
y_pred = df.loc[:, 'CLASS_PRED']
y = df.loc[:, 'CLASS']
# Examine the results
confusion_mat = confusion_matrix(y, y_pred)
confusion_matrix_df = pd.DataFrame(confusion_mat, columns=['predicted neg', 'predicted pos'])
matrix = np.array(['actual_neg','actual_pos'])
confusion_matrix_df = pd.DataFrame(confusion_mat,
columns=['predicted neg', 'predicted pos'])
confusion_matrix_df["results"] = matrix
df = confusion_matrix_df[['results','predicted neg','predicted pos']]
df['Total costs'] = None
total_cost = ida_cost(y,y_pred)
df.loc[df.index.max() + 1] = ['Total costs',None,None,total_cost]
return df
def run(ctx):
df = ctx.get_dataframe(num_rows='all')
results = evaluate(df)
ctx.emit(results)
/
Then run the following statement:
SELECT IDA_EVALUATE_RESULTS (CLASS_PRED, CLASS)
FROM IDA_TEST_PREDICTIONS;Python environment
from sklearn.metrics import confusion_matrix
import pandas as pd
# Define cost function from the problem description
def ida_cost(y, y_pred):
false_preds = y - y_pred
num_false_pos = (false_preds < 0).sum()
num_false_neg = (false_preds > 0).sum()
return 10 * num_false_pos + 500 * num_false_neg
# Get predicted and real class labels
test_preds = C.export_to_pandas("SELECT CLASS_PRED, CLASS FROM IDA_TEST_PREDICTIONS")
y_pred = test_preds.loc[:, 'CLASS_PRED']
y = test_preds.loc[:, 'CLASS']
# Examine the results
confusion_mat = confusion_matrix(y, y_pred)
confusion_matrix_df = pd.DataFrame(confusion_mat,
index=['Actual neg', 'Actual pos'],
columns=['Predicted neg', 'Predicted pos'])
print("Confusion matrix:\n", confusion_matrix_df)
print("\nTotal cost:", ida_cost(y, y_pred),"\n")Close the connection:
# close the connection
C.close()Step 7: Confusion matrix
The evaluation script should produce the following results:
| Results | Predicted neg | Predicted pos | Total costs |
|---|---|---|---|
| Actual neg: | 14326.0 | 1299.0 | |
| Actual pos: | 206.0 | 169.0 | |
| Total cost: | 115990.0 |
Using the confusion matrix and the total cost, we can now compare our model to others.
In this tutorial you learned how to evaluate a model using UDFs. If you want to learn more about how to create a machine learning model in an Exasol database, refer to the Jupyter Notebook section in our Data Science GitHub repository.
